We aim to use genetics and digital models to improve the sustainability of agricultural production. This Poster/Banner was made for the UK Dairy Day in Shropshire @UKDairyDay, which showcases the best breeding, husbandry, and technology for British dairy farming.
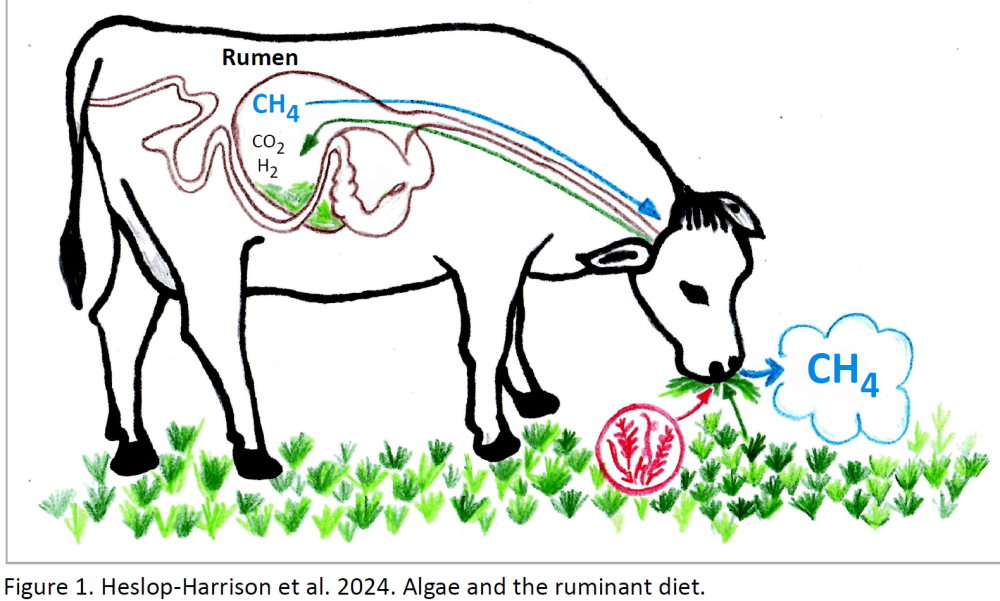
Cows do not directly use grass for growth: the microbes in their rumen produce enzymes that break down grass cell wall components like fibre and cellulose to a form that can be absorbed. Some products of the digestion are converted by the microbes to methane, a potent greenhouse gas. The genetics of the animal, the microbes in their rumen, and the feed all interact and contribute to methane production.
Our research We are an academic research group with long term objectives of increasing agricultural sustainability, ensuring food security, and improving livelihoods (profitability). We work on crop plants, including forage grasses, and farm animals at DNA, chromosome and genome levels. We use genomics, microscopy, bioinformatics, molecular genetics, and computer models/systems biology to gain a comprehensive understanding of fundamental processes related to genetics and breeding. Much work exploits genetics in wild relatives of domesticated species

Ruminant genomics We are working on projects on Bovidae evolution and biodiversity. Our work on the diversity of different cattle, sheep and goat breeds around the world, reveals new allelic diversity in growth related genes. We have shown how chromosomes evolve and change within breeds and between wild relatives, with a focus on major regions of the genome’s DNA that are highly repeated. Thus, research with collaborators around the world shows the patterns of evolution, recombination and fusion of repetitive DNA sequences, including the centromeres of chromosomes, ends (telomeres) and mobile, transposable elements including ERVs. Important work shows origin of new variation from structural (rather than DNA) changes in the genome. Overall, organization and chromosomal localization resolves events in bovid evolution and shows how the reservoir of genetic diversity can be exploited.

Grass genomics and agricultural greenhouse gases Cattle produce essential milk and meat from grass crops grown with low intensity in areas unsuitable for other crops. However, cows also produce methane, a potent greenhouse gas, through anaerobic microbial digestion of forages, and from nitrogen emissions, arguably causing as much greenhouse gas as a diesel van. Reduction of methane production is a major environmental target. Improved microbial composition and animal genetics is important. Our work with forage grass diversity shows ways to generate new varieties with increased digestibility of cell walls, and higher lipid contents, that can reduce individual animal methane emissions while minimizing concentrates.
Medium term applications We evaluate the environmental outcomes of the genetics of crops and animals, linked to agricultural practices and their consequences, including their economic and social impacts. Leveraging the insights from our research allows improving agricultural genetics, environment and mitigating climate change.
Self-learning digital twins for sustainable land management Joint research with Baihua Li’s team at Loughborough University project is developing an Artificial Intelligence algorithm (Self-Learning Digital Twin) for sustainable land management. Using computational modelling, and environmental measurements, the outcome will give environmental insights into use of genetics and husbandry, and help farmers reduce greenhouse gas emissions, saving feed and land resources. More generally, digital models will increasingly allow complex genetics to be exploited to reduce environmental impact and improving welfare, while continuing the exceptional trajectory of increasing food production while reducing costs, as in the last 50 years.
Academic publications from our group which relate to the work above (Publications page of http://www.molcyt.org will have links, or Google will find them):
Biscotti MA, Olmo E. & Heslop-Harrison JS 2015. Repetitive DNA in eukaryotic genomes. Chromosome Research 23: 415-420.
Li R, Gong M, Heslop-Harrison JS +31 2023. A sheep pangenome reveals structural variations and effects on phenotypes. Genome Research 33: 463-477.
Mustafa SI, Schwarzacher T & Heslop-Harrison JS 2022. The nature & chromosomal landscape of endogenous retroviruses (ERVs) in sheep. DNA 2: 86-103.
Mustafa SI, Heslop-Harrison JS & Schwarzacher T 2022. Mitochondrial genomes from Iraqi meriz goats. Iranian Applied Animal Science 12: 321-328
Escudeiro A, Ferreira D, Mendes-da-Silva A, Heslop-Harrison JS, Adega F & Chaves R 2019. Bovine satellite DNAs–a history of the evolution of complexity and its impact in the Bovidae family. European Zoological Journal 86: 20-37.
Masters LE, Tomaszewska P, Schwarzacher T, Zuntini AR, Heslop-Harrison P, Vorontsova MS 2023. Phylogenomic analysis reveals evolutionary origins of 5 independent clades of forage grasses within the African genus Urochloa. Annals of Botany 133:725-742
Hanley SJ, Pellny TK, de Vega JJ, Castiblanco V, Arango J, Eastmond PJ, Heslop-Harrison JS, Mitchell RA 2021. Allele mining in diverse accessions of tropical grasses to improve forage quality & reduce environmental impact. Annals of Botany 128: 627-637.
Tomaszewska P, Vorontsova MS, Renvoize SA, Ficinski SZ, Tohme J, Schwarzacher T, Castiblanco V, De Vega JJ, Mitchell RA & Heslop-Harrison JS 2023. Complex polyploid and hybrid species in an apomictic and sexual tropical forage grass group: genomic composition and evolution in Urochloa (Brachiaria) species. Annals of Botany 131: 87-108.
Tomaszewski C, Heslop-Harrison JS, Anhalt UC & Barth S 2010. Fine mapping of quantitative trait loci for biomass yield in perennial ryegrass. In Sustainable Use of Genetic Diversity in Forage and Turf Breeding (pp 461-464). Springer Netherlands.
Anhalt UCM, Heslop-Harrison JS, Piepho HP, Byrne S & Barth S 2009. Quantitative trait loci mapping for biomass yield traits in a Lolium inbred line derived F2 population. Euphytica 170: 99-107.
The Banner and Leaflet about this project can be downloaded here as PDFs.