304. Heslop-Harrison JS, Schwarzacher T. 2013. Nucleosomes and centromeric DNA packaging. Proc Nat Acad Sci USA 110 (50): 19974–19975. http://www.pnas.org/content/110/50/19974.full.pdf+html
http://dx.doi.org/10.1073/pnas.1319945110 –
Author manuscript: Author Version 2013 –
Powerpoint figure: Nucleosome Figure PNAS Schwarzacher HeslopHarrison
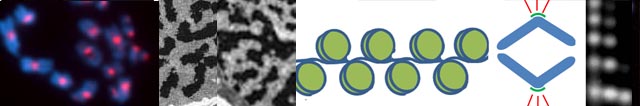
The eukaryotic chromosome is a conserved structure, with the DNA double-helix wrapping around octamers of histone proteins to form the chromatin which is further packaged into chromosomes. The centromere defines the kinetochore, the region of spindle microtubule attachment that pulls the two replicated chromatids of each chromosome apart during cell division (Fig. 1) leading to fidelity in transmission of genetic information. Like the telomere, centromeres are well defined morphologically and functionally, but their DNA sequence shows no conservation between species and their coiling into chromatin is still poorly understood. In PNAS, Zhang et al.(1) give new insight into the CenH3- nucleosomes associated with the rice CentO satellite sequence and adjacent regions of DNA having implications for centromere specification, activity and evolution.
In rice, Zhang et al. (1) have shown how ChIP-sequencing on a relatively large scale has revealed the phasing and nature of spacing of CentO and non-satellite, CenH3 -nucleosomes. This has made a significant advance in our understanding of the DNA:histone variant interaction and the nature of centromeric chromatin packaging, crucial to chromosome structure and the specification of centromeres, assigning a functional role to the abundant but enigmatic class of centromeric satellite DNA. It will now be important to integrate these results with a dynamic picture of cenH3 loading (15). New, super-resolution microscopy methods (16), careful assembly of satellite DNA arrays (9, 17), and comparative studies of different organisms will help with answering the long-standing questions of what defines a centromere. How, and when, is the DNA underlying the centromeric nucleosomes defined in the cell? Why, and how, does this DNA become the target to associate with the centromeric histone variant? These large questions must be partitioned to tractable experiments, and it needs to be considered whether imaging, mutant, sequencing/informatics or biochemical approaches are going to be most informative. Exquisite control of centromere function, and the highest stability of the process, is essential for correct chromosome segregation. This requires both robust and redundant control, includes evolutionary selection for both satellite and other DNA sequences with their interacting counterparts. Application of knowledge of centromere specification and control has potential not only for understanding aneuploidy-related diseases, but also may be exploited in generation of new hybrid plant varieties.
Original Powerpoint Artwork Nucleosome Figure PNAS Schwarzacher HeslopHarrison CC-BY-SA
Author version of manuscript Nucleosomes and Centromeres PNAS Heslop-Harrison Schwarzacher Author Version 2013

Congratulations sir
Sent from Samsung Mobile